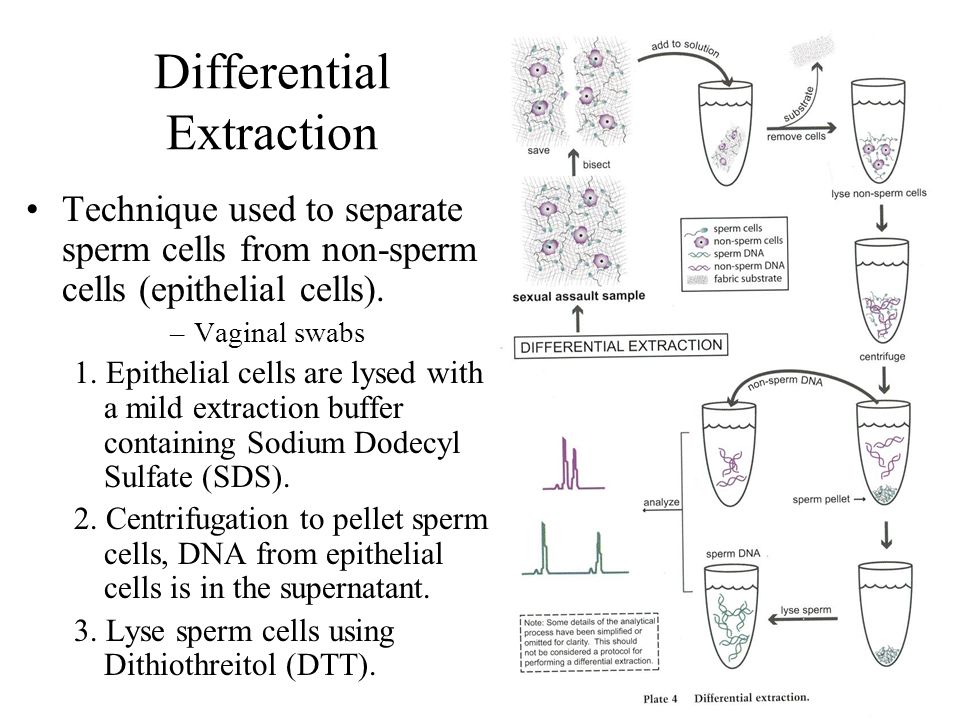
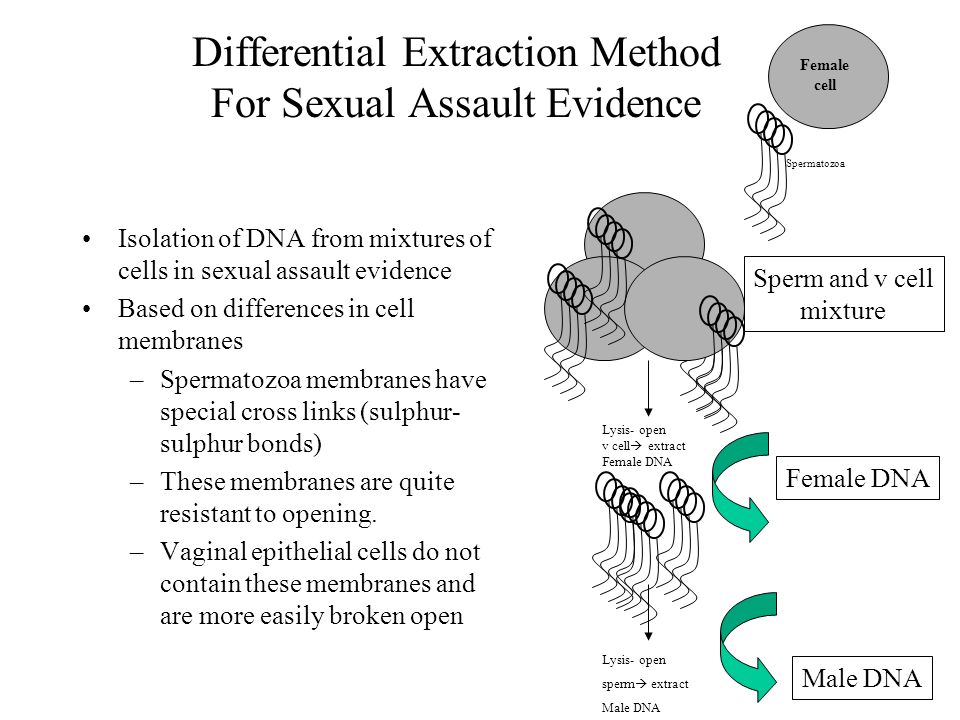
Isolation Of Dna From Fish Sperm

🛑 ALL INFORMATION CLICK HERE 👈🏻👈🏻👈🏻
Isolation Of Dna From Fish Sperm
Deoxyribonucleic acid (DNA) is a complex nucleic acid containing the genetic code with the instructions for the development and functioning of all known living organisms, with the exception of some viruses. The DNA segments that carry this genetic information are called genes. DNA is transcribed into RNA which is then used as a template in the synthesis of proteins.
DNA isolation refers to the process of extracting DNA from a cell in a pure form. The extraction of DNA is an important preliminary step in which purified DNA is obtained from other cellular components such as proteins, RNA and lipids. DNA can be isolated from any nucleated cell from diverse sources, both living and dead, such as whole blood, hair, sperm, bones, nails, tissues, faeces, shed feathers, egg shells, saliva, epithelial cells, urine, bacteria, animal tissues or plants.
Isolation of DNA is required for a variety of applications in science, medicine and forensics. For example, scientists introduce DNA into the cells of animals or plants for SNP analysis, DNA methylation analysis, copy number variation (CNV) or comparative genomic hybridization (CGH) analysis, Southern Blotting, PCR etc. In medicine, DNA isolation is helpful for diagnostic purposes. Furthermore, DNA extraction is an important tool in forensic science for the identification of individuals (for example rapists, thieves, accident, or war victims), paternity determination, body identification etc.
There are a number of different procedures for the preparation of genomic DNA. They all start with some form of cell lysis, followed by deproteination and recovery of DNA. The main differences between various approaches lie in the extent of deproteination and in molecular weight of the DNA produced. Factors affecting the methods of DNA isolation are the age,source,and size of the sample.
The presence of proteins, lipids, polysaccharides etc. during DNA preparation can interfere with DNA analysis methods by reducing the quality of DNA. The extraction methods to efficiently purify DNA from various sources have to be adapted depending on factors such as sample size, the freshness of the sample, and the biochemical content of the cells from which DNA is being extracted. The isolation method must vary depending on the size of sample. The freshness of the sample also affects the extraction technique. Extraction methods are also variable according to the biochemical content of the source cells. For example, in the case of bacteria, the main biochemicals present in a cell extract are protein, DNA and RNA. Therefore, phenol extraction or protease treatment, followed by removal of RNA with ribonuclease, leaves a pure DNA sample. These treatments may not be sufficient if the cells also contain significant quantities of other biochemicals. Plant tissues are particularly difficult in this respect as they often contain large amounts of carbohydrates that are not removed by phenol extraction.
DNA molecule
Fish fins are an excellent and reliable source of high quality DNA with a number of advantages. In most cases DNA is obtained from the collection of blood from the fish. In the case of rare or endangered species, this method is not advisable. Therefore, fin tissue is often used for DNA extraction because sampling is relatively fast, logistically simple, and is non-lethal. A small piece of tissue provides plenty of DNA for PCR and restriction digests. Most importantly, collection of fish fin sample does not harm the fish. This is a very important consideration especially relevant regarding rare and the ornamental species. Fish fin samples are used for DNA-based studies on genetic diversity, mating systems and parentage determination of fish populations with minimal disturbance.
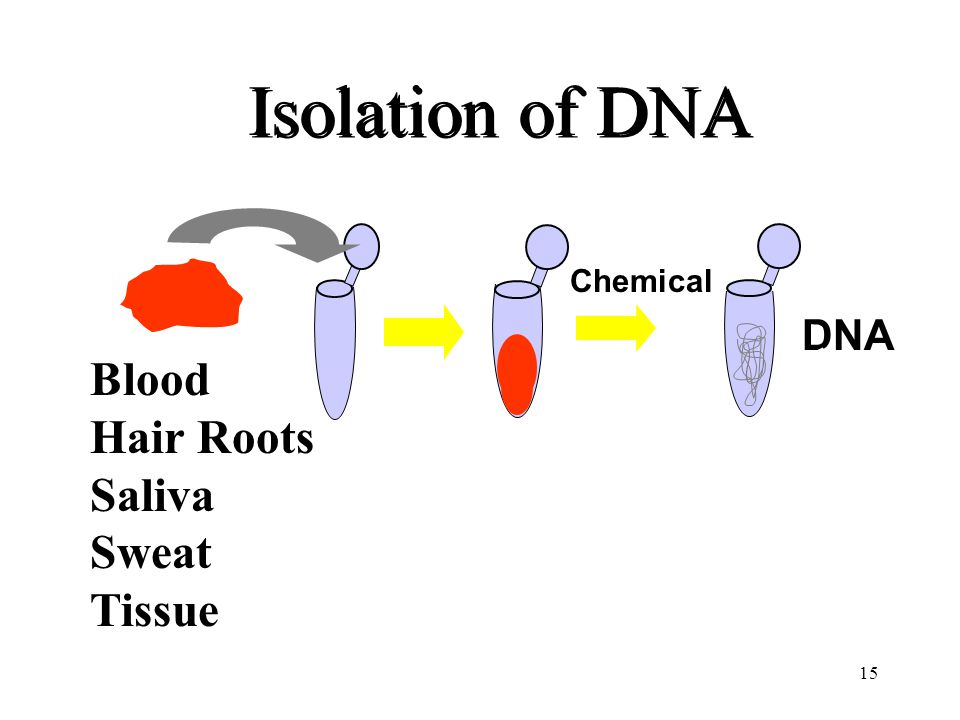
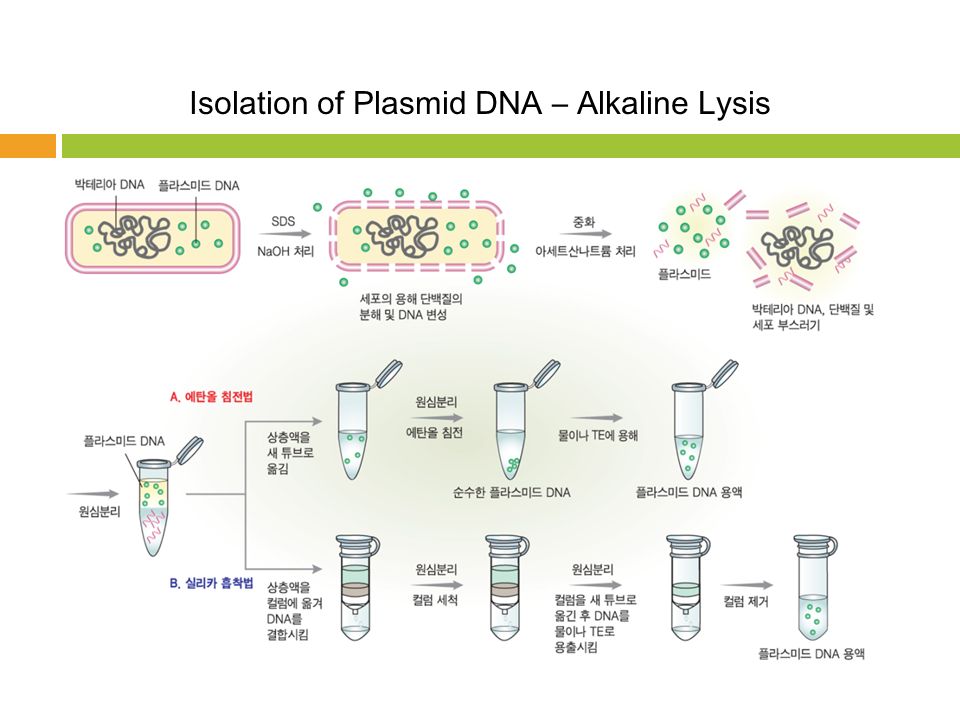
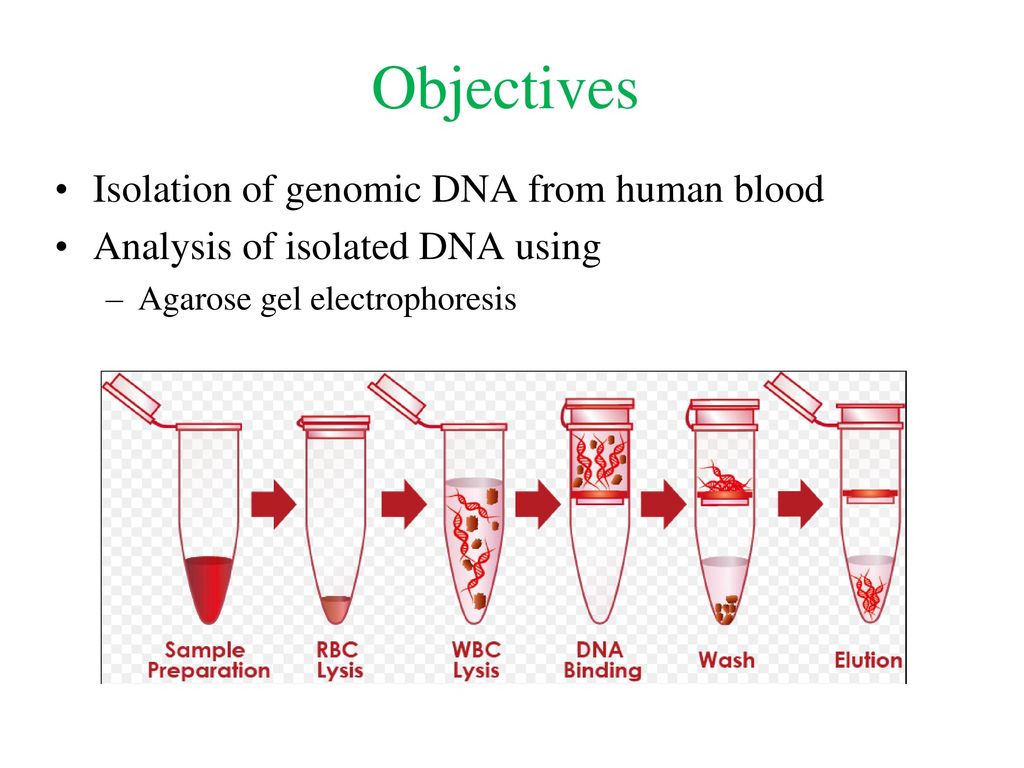
The isolation of DNA usually begins with the lysis of cells or tissues inorder to destroy the protein structures and allows the release of nucleic acids from the nucleus. Lysis is carried out in a lysis solution containing important ingredients: sodium chloride which provides an osmotic shock to the cells; Tris HCl, which is a buffer to retain constant pH; EDTA, which sequesters the divalent metal ions that is required for nuclease activity and thereby inhibiting its action; a detergent, usually SDS, which disrupts the cell membrane and nuclear envelope, causing the cells to burst open and release their DNA. The DNA is still rapped very tightly around histone proteins. Proteinase K (a serine protease) is the common enzyme used in DNA extraction that cuts apart the histones to free the DNA and finally results in the breakdown of cells and dissolving of membranes.
The nucleic acids are then purified from the protein- nucleic acid complex by phase extraction with a mixture of organic solvents namely Phenol, Chloroform and Isoamyl alcohol in a ratio of 25:24:1. Phenol dissociates proteins from DNA. Chloroform denatures the proteins and lipids and helps to maintain the separation of the organic and aqueous phase. It also makes the DNA less soluble in the phenol, thus reducing losses to the organic phase. Isoamyl alcohol is often added to prevent foaming. At pH 7-8, the DNA partitions to the aqueous phase while the protein is denatured and extracted into water- immiscible organic phase, which is separated from the nucleic acid containing aqueous phase by centrifugation. When large amount of protein is present, it forms a white precipitate between the organic and aqueous phase.
The DNA is then precipitated with cold ethanol or isopropanol after adjustment with 3M sodium acetate and then centrifuging. The DNA is insoluble in the alcohol and will come out of solution, and the alcohol serves as a wash to remove the residual salts. The alcohol is then removed, and DNA is stored in a biological buffer, like TE (Tris-EDTA) buffer. Contaminating RNA in the DNA sample can eliminated by digestion with an RNase.
However, since shearing forces are generated at every step, the resulting DNA molecules in the final preparation rarely exceed 100-150 kb in length. DNA of this size is adequate for Southern analysis on standard agarose gels, as a template in PCRs, and for the construction of genomic DNA libraries in bacteriophage λ vectors.
Copyright @ 2021 Under the NME ICT initiative of MHRD
Extraction of DNA from fish blood and sperm
Extraction of DNA from Fish Fins (Theory) : Molecular Biology Virtual...
(PDF) An efficient method of dna extraction from fish fin
Dna Isolation Methods | Encyclopedia.com
Isolation Of Dna From Fish Sperm | ВКонтакте
A, B. Representative (1%) gel of DNA isolated by modified salt extraction method from 14 different species of the fish. (M) DNA ladder mix (SM 331, Fermentas), (1) L. rohita, (2) L. calbasu, (3) L. gonius, (4) C. catla, (5) C. mrigala, (6) C. reba, (7) S. seenghala, (8) R. rita, (9) B. bagarius, (10) W. attu, (11) O. pabda, (12) O.mossambicus, (13) M. armatus, (14) C. fasciata (15) S. sarana, (16) C. idella, (17) H. molitrix, (18) C. carpio, (19) O. niloticus, (20) O. aureus, (21) M. bleekeri, (22) M. vitattis, (23) E. vacha, (24) C. garua, (25) C. punctata, (26) C. marulius, (27) C. chitala, (28) G. chapra and (29) N. notopterus
Representative (1%) agarose gel electrophoresis of isolated mitochondrial DNA by modified salt extraction method by with CO1 gene specific marker. (M) DNA ladder mix. (SM 331, Fermentas), (1) E. vacha, (2) C. garua, (3) W. attu, (4) O. pabda, (5) M.armatus, (6) N. notopterus and (7) control without DNA template.
Comparison of DNA concentration, yield and purity isolated from 29 freshwater species of fish.
One way ANOVA table of isolated DNA quantity of 29 fish species.
Content may be subject to copyright.
Content may be subject to copyright.
Deoxyribonucleic acid (DNA) n ot only stores hereditary
information’s but transfers these info rmation’s generation
after generation from parents to their offspring’s. It is known
as h ereditary material in all living organisms and massively
used in various molecular studies. The principle and isolatio n
techniques play important r ole for succ essful extraction of
purified and c onsiderable qu antity of DNA bec ause cell is t he
complex of different organelles and biolo gical molecule s.
Scientifically DNA is used in diagnostic purposes , forensic,
genetic, and medical studies. It is also use d to manipulate
bacterial, plants and animals cells as well as for pathogenic,
paternity and organismic identificatio n. (Srividya et al .,
2011). Contaminants in isolated DNA like lipids , proteins,
polysaccharides, different inorganic an d organic compounds
interfere with its further analysis especially by Polymerase
Chain Reaction and decrease the quality and storage life of
DNA (Bauer and P atzelt, 2003). The extent o f DNA isolation
and purity depe nds upon many factors including sa mple size,
extraction methods and storage of sample.
In the rece nt years iso lation of D NA from the sources that are
not destructive for th e organism is th e most interesting and
emerging methodology . This t echnique is extremely
important for DNA isolation from the endangered or
threatened spec ies to study their co nservation, population,
diversity and genetic asses sment. DNA isolation can be do ne
from eggshells, feces, sloughed off skin of whale and snake,
urine, feathers, hairs and bones i n feces pellets of carnivores.
These sources provide poor and low quantity DNA thus
hardly used for individual i dentification. On the other hand
blood, fins, scales, skin an d muscles can be used for
successful isolation of good quantity and quality DNA
without any po tential damage to the animals. The isolated
DNA can be used to determine genetic polymorphism
between and within the populatio ns, reconstruction of
pedigree, related estimates, sex determination and individual
identification. In case of fish isolation of DNA from scales
and fins is desirable be cause both tissue s are attractiv e source
of DNA isolation an d proved to be non -destructive for fish
(Wasko et al ., 2003; Ku mar et al ., 2007). F ins a re given
preference over scales for DNA isolation because scales ar e
not present in all species of fish. Many researchers (Bruyn et
al , 2011; Raja et al , 2011) tried to iso late DN A from th e
animals preserved in the natural history museums but t hey
were n ot successful to r ecover intact and sufficient quantity
of DNA from the preserved specimens. Prob ably these
organisms were collected before advancement in molecular
Pak. J. Agri. Sci ., Vol. 53(4), 843-850 ; 2016
ISSN (Print) 0552 -9034, ISSN (O nline) 2076-0906
AN EFFICIENT M ETHOD FOR DNA ISOLATION FROM FISH FIN
Haji Muhammad 1,2 , Zafar Iqbal 1 , Muhammad Usman Iqbal 2 , Tayyaba Younas 2 and Qamar
1 Department of the Zo ology, University o f the Punjab Laho re, Pakistan; 2 School o f Biological Sciences, Univers ity
* Corresponding au thor’s e-mail: qamar.sbs@p u.edu.pk
Genomic DNA isolation is fu ndamental step to study genetic diversity, p hylogeny of specie s and cloning of de sired genes. The
preferable protocol needs to be efficient, less time consuming, economic and non-destructive particularly for the endangered
species. The cu rrent study was aimed at eco nomic isolation o f considerable quantity of DNA ma nually from a p articular ti ssue
of the fish without any potential damage to the animal. For this purpose fin tissue was prefe rably selected over scale due to its
universal oc currence in fish. A total of 180 sample s of fin tiss ues belo nging to twenty nine species of f resh wate r fish preserved
in different preservativ es, were used fo r DNA isolation. DN A could not be isolated from the fin tissues preserved in fo rmalin,
alcohol, air dried and ice preserved fins more t han one week by any of t wo previously described methods including salt
extraction and u rea treatme nt. However, i ntroducing some modific ations in salt ext raction method resulted in large qua ntity of
high quality DNA from those fin tissues that were isolated from the living animal and immediately pr eserved in absolute
ethanol. The modifications include use of low quantity of proteinase K and slight increase in incubation period due to hard
nature of fin tissue. The extracted DNA by the modified salt extractio n method was very pure, of high quality and resulted in
successful PCR amplific ation by Random Amp lified Polymorphic DN A primer and cy tochrome oxidase sub unit 1 gen e
specific primers. This modif ied procedure results in highest y ield of isolated DNA reported so far. The method is particularly
suited f or DNA isolatio n f rom enda ngered and spa rse spe cies as it r equires minute quantity of fin t issue, w ithout any
Keywords: DNA, fis h fin, salt extraction method, RAPD, CO1
Although much biological research depends upon species diagnoses, taxonomic expertise is collapsing. We are convinced that the sole prospect for a sustainable identification capability lies in the construction of systems that employ DNA sequences as taxon 'barcodes'. We establish that the mitochondrial gene cytochrome c oxidase I (COI) can serve as the core of a global bioidentification system for animals. First, we demonstrate that COI profiles, derived from the low-density sampling of higher taxonomic categories, ordinarily assign newly analysed taxa to the appropriate phylum or order. Second, we demonstrate that species-level assignments can be obtained by creating comprehensive COI profiles. A model COI profile, based upon the analysis of a single individual from each of 200 closely allied species of lepidopterans, was 100% successful in correctly identifying subsequent specimens. When fully developed, a COI identification system will provide a reliable, cost-effective and accessible solution to the current problem of species identification. Its assembly will also generate important new insights into the diversification of life and the rules of molecular evolution.
Two simple and rapid procedures of DNA extraction based on Chelex resin were compared and optimized to get reliable amplification by polymerase chain reaction (PCR) from diverse sources of fish tissue (fin, liver, muscle, blood, scales, and individual eggs) at numerous nuclear loci (13 microsatellites and 4 lacZ transgenes) and mitochondrial loci (ND5/6 and cytochrome b/D loop). The first procedure worked for the majority of markers under investigation. However, it did not result in satisfactory PCR products for six microsatellite loci and for one mitochondrial DNA domain. This problem was addressed with the second procedure by adding proteinase K from the extraction start. This pretreatment provided an all-or-none improvement, as it had a strong positive effect on ''refractory'' loci (much better PCR yield and reduced number of unspecific and supernumerous bands) but did not quantitatively enhance or attenuate the amplification of all other loci.
A specific DNA extraction method for sea anemones is described in which extraction of total DNA from eight species of sea anemones and one species of corallimorpharian was achieved by changing the standard extraction protocols. DNA extraction from sea anemone tissue is made more difficult both by the tissue consistency and the presence of symbiotic zooxanthellae. The technique described here is an efficient way to avoid problems of DNA contamination and obtain large amounts of purified and integral DNA which can be used in different kinds of molecular analyses.
We evaluated two methods, boiling and microwave irradiation, for the rapid isolation of DNA from barbel, adipose fin, and blood of channel catfish Ictalurus punctatus. Compared with routine DNA isolation methods, these procedures were fast (microwaving, 2–4 min; boiling, 10–18 min), simple, and inexpensive (about US$0.10/fish). Samples of DNA isolated from barbels of small (mean ± SD, 2.9 ± 1.0 g) and large (290 ± 43 g) fish were of high purity (ratios of absorbances at 260 and 280 nm, A260/A280 = 1.72–1.90) and were between 0.29 and 0.67 μg/μL in concentration. Samples of DNA isolated from barbel, adipose fin, and blood of small fish, by either method, were used successfully for analysis by polymerase chain reaction (PCR). Samples isolated by boiling of barbel and blood from large fish also proved useful for PCR analysis. These DNA isolation procedures would be useful for rapid genetic screening of channel catfish. Removal of the barbel for tissue analysis would also enable direct marking of fish, and after analysis, individuals designated for further study could be identified.
Collections of old scales from salmonid fishes can be found in many fisheries institutions in Europe and North America. Such scales have been shown to be useful as a source of DNA, which can be used for analysis of microsatellites. We here describe the technical procedures in relation to extraction and PCR amplification of DNA from old scales. Further, we describe case stories of the application of old scale DNA analysis for conservation genetics with regard to: 1) Identification of native populations, 2) assessment of loss of genetic variation, 3) estimation of effective population size, 4) evaluation of anthropogenic effects on the genetic population structure/mode of migration and 5) the temporal stability of the genetic population structure. Finally, we describe the perspectives for future genetic studies using old scale DNA. In particular, we evaluate other types of genetic markers such as mtDNA and loci subjected to selection, in association with old scale DNA, as tools for prioritising populations for conservation.
This study describes the comparison of five DNA extraction methods (urea-SDS-proteinase K; phenol-chloroform; salt extraction; SureFood® PREP Allergen kit and Wizard® Genomic DNA Purification kit) in terms of their ability to extract high yields of pure, readily amplifiable DNA from the muscle tissue of 29 fish species available in South Africa. Although variations in the yield and purity of extracted DNA were observed between methods, all five methods produced DNA suitable for PCR amplification. Overall, the SureFood® PREP method was of the simplest and least hazardous to perform, extracting significantly (P < 0.05) higher DNA yields than all other methods evaluated.
A protocol used routinely for rapid ancient DNA extraction was applied to fish tissue archived over 80 years ago. The method proved successful, whereas other extraction protocols failed. Researchers working on DNA from older archived fish samples are encouraged to continue to concentrate their efforts on 'white-eye' specimens, which indicate an alcohol-based fixative and are thus likely to yield viable DNA. (C) 2011 The Authors Journal of Fish Biology (C) 2011 The Fisheries Society of the British Isles
Discover more about: DNA Extraction
September 2012 · Biochemical Genetics
November 2016 · Pakistan Journal of Agricultural Sciences
Genomic DNA isolation is fundamental step to study genetic diversity, phylogeny of species and cloning of desired genes. The preferable protocol needs to be efficient, less time consuming, economic and non-destructive particularly for the endangered species. The current study was aimed at economic isolation of considerable quantity of DNA manually from a particular tissue of the fish without any ... [Show full abstract] potential damage to the animal. For this purpose fin tissue was preferably selected over scale due to its universal occurrence in fish. A total of 180 samples of fin tissues belonging to twenty nine species of fresh water fish preserved in different preservatives, were used for DNA isolation. DNA could not be isolated from the fin tissues preserved in formalin, alcohol, air dried and ice preserved fins more than one week by any of two previously described methods including salt extraction and urea treatment. However, introducing some modifications in salt extraction method resulted in large quantity of high quality DNA from those fin tissues that were isolated from the living animal and immediately preserved in absolute ethanol. The modifications include use of low quantity of proteinase K and slight increase in incubation period due to hard nature of fin tissue. The extracted DNA by the modified salt extraction method was very pure, of high quality and resulted in successful PCR amplification by Random Amplified Polymorphic DNA primer and cytochrome oxidase subunit 1 gene specific primers. This modified procedure results in highest yield of isolated DNA reported so far. The method is particularly suited for DNA isolation from endangered and sparse species as it requires minute quantity of fin tissue, without any detrimental effect on fish.
Keywords: DNA, fish fin, salt extraction method, RAPD, CO1
March 2020 · International Journal of GEOMATE
April 2014 · Food Analytical Methods
DNA barcoding is a sequencing-based method that can be used for the identification of fish species in a regulatory setting. The objective of this study was to compare modified versions of three DNA extraction kits (i.e., Qiagen DNeasy Blood and Tissue Kit, Sigma-Aldrich Extract-N-Amp Kit; and Life Technologies MagMax-96 DNA Multi-Sample Kit) and two PCR setup methods (manual vs. automated) for ... [Show full abstract] use in DNA barcoding, with a focus on minimizing time, costs, and labor. DNA was extracted from 83 fish products using each of the three kits and the results were compared based on sequencing success and sequencing quality parameters. A subset of 14 fish products was also tested in triplicate to compare PCR setup methods. Initially, reduced sequencing success was observed with the MagMax Kit (88%) compared to the other two kits (95-96%); however, after PCR and sequencing were repeated for DNA samples that initially failed, all three methods showed very high sequencing success (98-99%). Overall, the modified Extract-N-Amp Kit offered the greatest reduction in time and costs, while the DNeasy Blood and Tissue Kit produced sequences with the highest quality and highest initial success rates. Automation of the PCR setup process resulted in slightly greater success (100%) compared to manual PCR setup (98%), and reduced the potential for human error that may result from manual pipetting. The results of this study demonstrate the advantages of incorporating rapid and/or automated methods into the DNA barcoding workflow, especially with regard to high-throughput operations.
September 2018 · Zoology and Ecology
The present study was aimed to DNA extraction and PCR amplification of mitochondrial DNA of a varied size range using the non-invasive samples of fish. As many obstacles arise when obtaining an adequate amount of DNA from samples of endangered species, using non-invasive methods of sample collection and DNA extraction should be developed. To this end, we tested and standardised the reliable ... [Show full abstract] methods based on DNA extraction from a single scale of small Cyprinidae fish Pethia conchonius. In total, 13 specimens were used for DNA extraction, including 7 single scales and multiple scales. The DNA fragments of different length representing four mitochondrial fragments of different genes were amplified with a variable success after DNA extracted from a single scale, multiple scales or fish clips was used in PCR reactions. All examined DNA samples extracted from a single scale provided 100% amplification success of up to 381 base pairs fragments. Methods described here have a great applicability in DNA extraction from the small size fish and could be useful in conservation genetics as well as in wildlife forensics.
© 2008-2021 ResearchGate GmbH. All rights reserved.
Vintage Nudist Pictures
Chubby Riding Porn
Naked Girl Home
Porno Kids Sperm
Mature Missionary Xxx