176 Questions for Scientists and the WHO (Part 2)
Decentralised Radical Autonomous Search Team Investigation into Covid-19 (DRASTIC)This list of questions has been generated by a group of researchers working together on Twitter to investigate the origins of SARS-COV-2 and explore the possibility that it resulted from a laboratory acquired infection among other theories.
The researchers are microbiologists, renegade virologists, investigative reporters, analysts and ordinary citizens, some of whom prefer to remain anonymous in order to protect their positions, privacy and safety.
There are some repeat questions as it is a work in progress!
Any questions should be addressed to the curator of this list: billybostickson@gmail.com
or via Twitter: @billybostickson
Questions are credited via Twitter handles.
Questions 86 to 176
Via @MonaRahalkar
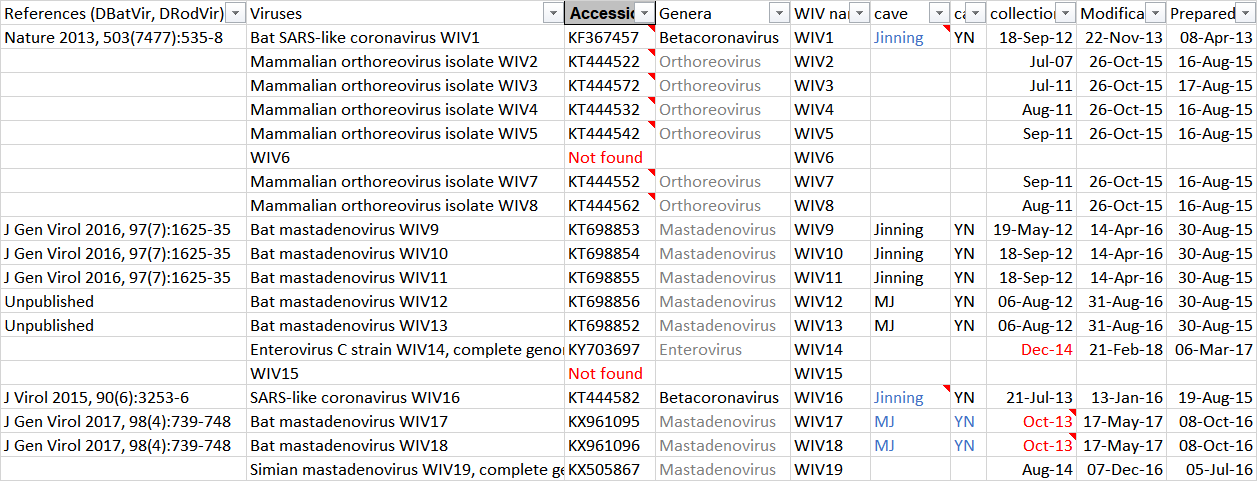
86. Were the faecal swab samples of RatG13 in the WIV Lab Freezer discarded few months ago?
87. Is RaTG13 and 4991 really the same? Did WIV isolate the RatG13 virus from 4991?
88. Why did Shi avoid referring to the Ge et al 2016 ref when writing her paper on the new pneumonia outbreak in 2020, and is she aware if the Li Xu master’s thesis findings?
89. If the Eco Health foundation is there to update people with important information regarding zoonotic spill over, why wasn’t the 2012 pneumonia outbreak mentioned by any of the programmes like Predict, Eco health and WHO?
90. How many people visited the Mojiang mineshaft in Yunnan up to now, and is it still open to researchers?
91. Did any of the bat researchers become sick recently during August to September 2019 or earlier?
92. Did the WCDC or other institutes in Wuhan have live bats and use them in experiments?
93. Do any other institutes in Wuhan, not only WIV, have samples from Mojiang Yunnan mineshaft?
94. Were any of the lab animals infected with the bat guano from the Mojiang bats? If so which animals?
98. Why is the Master and PhD thesis study of the Mojiang miner’s Pneumonia deaths not mentioned anywhere?
Images of the Thesis:





99. If the Mojiang pneumonia cases were known to Zhong Nanshan, Zhengli Shi, George Gao Fu (Now Head of CCDC) and QinJin of IPB (Beijing), why did none of them, report it to the WHO or discuss it in their subsequently published papers?
100. If there is nothing to hide about the SARS like pneumonia outbreak in Mojiang:
A. Why was this story told in a different way by Zhengli Shi in recent media statements, for example:
“Fungus causing pneumonia”
B. Why was the reference to a SARS CoV being a cause of the Mojiang pneumonia outbreak in 2012 never mentioned in English Language Papers, News Reports or EcoHealthAlliance reports, and only mentioned in the Chinese thesis?
@flavinkins
101. What was the isolate WIV15?
102. Have all the possible viruses causing the Miner pneumonia in Mojiang been isolated and why did the Miners have positive results for SARS antibodies?
103. What kind of animals and what type of experiments were done in the Hubei CDC labs?
104. Why has the exact, detailed list of all WIV samples, isolates, etc. as well as the Wuhan University and the Hubei CDC samples and isolates not been shared with the WHO or other countries?
105. Are there any humanized mice currently at WIV Labs and what parts of the mice were humanized?

106. Do WIV, WCDC or Wuhan University “Institute of Model Animals” use rabbits, monkeys, ferrets, tree shrews, golden hamsters or other NHP animals for experimenting with live viruses?

107. Are there any Bat cell lines maintained in the WIV Lab or other Wuhan Labs and for what purposes?
108. Are there any experiments going on for insertion of Furin Cleavage Sites (FCS) into different coronaviruses similar to the infectious bronchitis virus experiments by Cheng et al (2019)?
Reference: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6832359/?
109. What was the content of the previously online but recently deleted 61.5 MB SQL virus database administered by Professor Zhengli Shi? Has she made the password protected section with unpublished viral genome sequences available to the WHO or international scientists?

110. Are there records of freezer, lab equipment or other equipment movement that could indicate the recent removal and destruction of samples?
Apart from the disposal of scientific research and office equipment from the WIV Xiaohongshan Park and Zhengdian Park branches announced on 8 January 2020:
https://web.archive.org/web/20200206111237/http://www.whiov.ac.cn/tzgg_105342/202001/t20200108_5484213.html
with contract awarded on 14 January 2020:
https://web.archive.org/web/20200206111004/http://www.whiov.ac.cn/tzgg_105342/202001/t20200115_5487914.html
111. Is there excess capacity in the sample storage facility or forensic evidence thereof indicating an extended use of said capacity when it was discovered to be empty at the time of investigation?
112. Why has an exact list of all experiments, isolates and samples in all Biolabs in Wuhan not been made available to international scientists or the WHO?
113. Did the WIV receive any virus samples from Guangdong Institute of Applied Biological Resources and the National Gene Bank for experimentation?
If so,
A. When?
B. For what purposes?
114. Why there were 3 different versions of RaTG13 all disagreeing with each other?
A. Ampliseq
B. Genbank
C. NGS contigs
Reference: https://drive.google.com/file/d/1ku1Sf2SlqO9Pbw-HzZNiqzPXy1g6nlUm/view?usp=drivesdk
115. Why does the mNGS data for RaTG13 contain nearly entirely Repeat sequences and unrecognizable reads (not resembling anything) with no bacterial reads?
@interne41914499
116. Can WIV coherently explain the differences in published sequence data for the RATg13 faecal swabs?
References for The RaTG13 "swabs": https://trace.ncbi.nlm.nih.gov/Traces/sra/?run=SRR11085797
An actual WIV swab:
https://trace.ncbi.nlm.nih.gov/Traces/sra/?run=SRR11085736
WIV bat and mouse tissue samples: https://ncbi.nlm.nih.gov/biosample?Db=biosample&DbFrom=bioproject&Cmd=Link&LinkName=bioproject_biosample&LinkReadableName=BioSample&ordinalpos=1&IdsFromResult=393936
infected with betacoronaviruses: https://trace.ncbi.nlm.nih.gov/Traces/sra/?run=SRR5819071

@flavinkins
117. Why are the Bacteria levels found in the RaTg13 faecal swab unusually low?
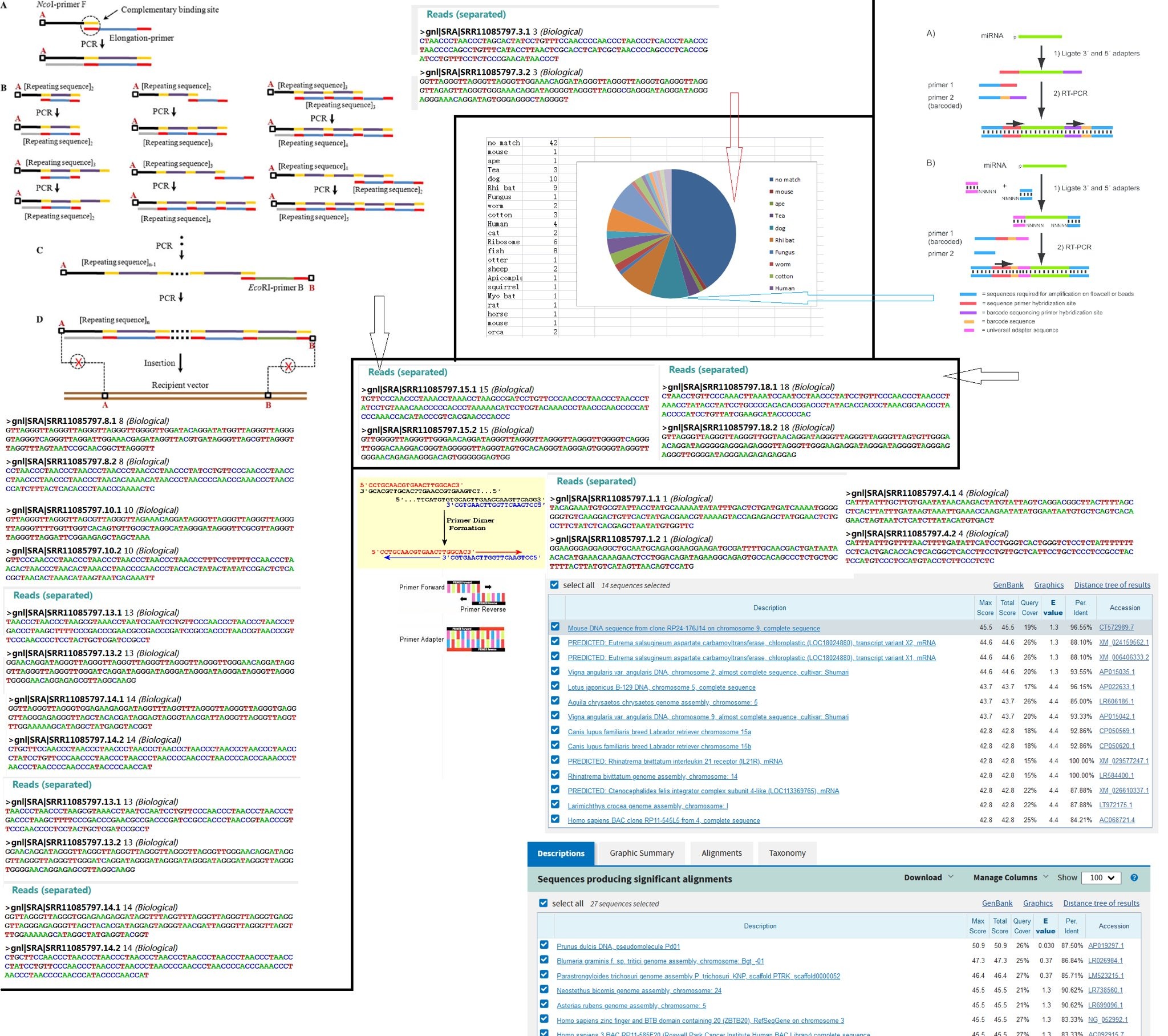
@billybostickson
118. Has anyone apart from WIV tried to amplify BatCov truncated sequence 4991 to generate a total genome sequence?
119. Has WIV shared the BatCov 4991 faecal swab with anyone else yet?
@JohnGal52793009
120. If the international groups (PREDICT, ONEHEALTH PLATFORM, ECOHEALTHALLIANCE ) are working with WIV to predict and prevent the next pandemic, how is it possible that they missed it, given that the closest known match in the WIV Lab freezer was collected under their project and guidance?
@interne41914499
121. What were the precise movements in late 2019 of WIV staff?
Did any isolate themselves for 14 days as Tian Junhua did years before when he was urinated on by bats in Yunnan?
http://xinhuanet.com//local/2017-05/03/c_1120909064_2.htm
and more recently:

122. What is the precise motivation behind the WIV taking databases associated with the program that identified RaTG13 offline and deleting traces of them?
123. Was there any official guidance similar to the 13 November 2019 instructions (http://archive.is/xOcDH#selection-809.2-817.178) issued to hospitals to look for COVID-19 symptoms in respiratory disease patients?
124. Was recent Chinese biosecurity legislation and regulations drafted as a reaction to laboratory leak spill over events? If so, what ones and where?
125. What type of viral pathogen research was being carried out by the WIV based on analysis of the now offline database until the early hours of 12 September 2019?
A. the types of files downloaded (http://archive.is/33Wlm)
B. WIV lab research direction in 2019
http://archive.is/bdJ0U#selection-3745.800-3745.1224
126. Which scientific teams were accessing the now deleted WIV database in 2019 according to and movements of researchers matched to database metadata records of:
Networks (http://archive.is/1uHeM) which accessed them
Countries (http://archive.is/BvH08)
Cities (http://archive.is/hyplV)
Provinces (http://archive.is/hypl)
127. What work was done on SARS-related coronaviruses done at the WIV:
ABSL-2 facility http://archive.is/zndQH#selection-1985.41-1991.45
BSL-2 http://archive.is/Fhvy9#selection-989.124-989.170
BSL-3 http://archive.is/Fhvy9#selection-989.124-989.170
BSL-4 http://archive.is/Fhvy9#selection-989.124-989.170
and other facilities at Zhengdian Scientific Park and its BSL-3 facility http://archive.is/V3GHk#selection-1549.0-1549.93 at Xiaohongshan Park,
as well as at Huazhong University, WHCDC and Wuhan University IMA ABSL3 Lab?
@flavinkins
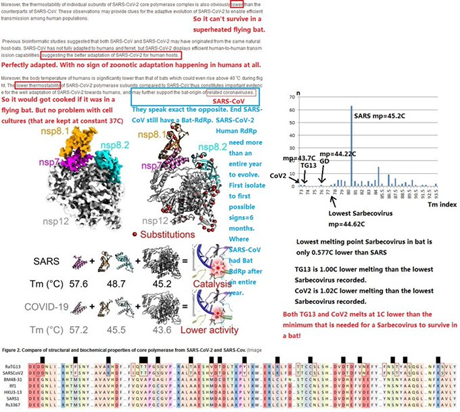
128. It has been claimed by many scientists, including Peter Daszak recently, that the most likely hypothesis is that SARS-COV-2 emerged from bats in South China, on the border area with Myanmar. How do you explain that SARS-COV-2 could not even survive in a live bat, due to the high internal body temperatures generated by Ra type bats during flight?
Reference: Image
129. What is RaTG13 and why there were 3 different versions of its sequences which all disagree with each other?
A. Ampliseq
B. Genbank
C. mNGS contigs
Reference: https://drive.google.com/file/d/1ku1Sf2SlqO9Pbw-HzZNiqzPXy1g6nlUm/view?usp=drivesdk
130. Why does the mNGS data for the faecal sample sequence of RatG13 contain nearly entirely repeat sequences and unrecognizable reads (not resembling anything) with no bacterial reads?
131. How do you explain the high level of “garbage sequences” which indicate a BatCov/4991 faecal sample with nearly no RNA in it—template free?
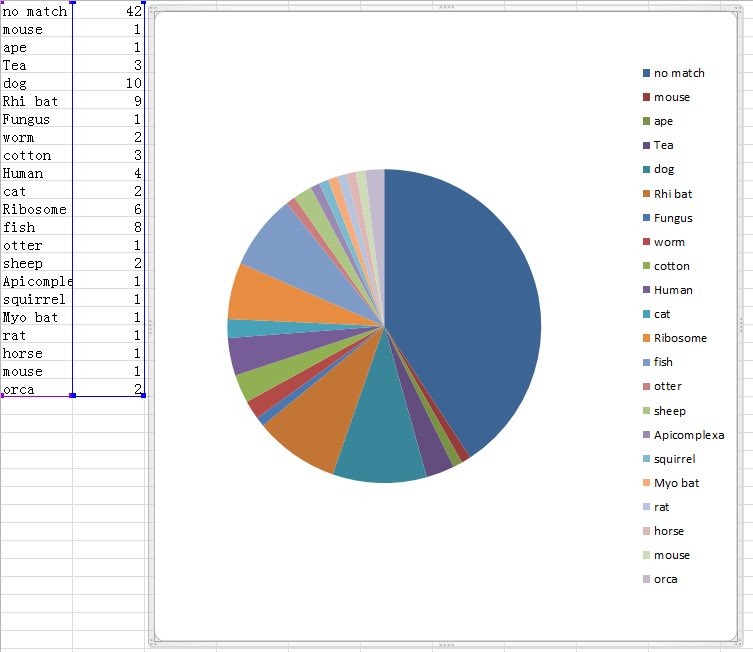
131. How could the ~252Kb virus reads (which happen to be the only Riboviria in there) be even there at high quality—if nothing else were there?
132. Does WIV admit that their bat faecal swab containing BatCov/4991 from which RaTG13 was allegedly finally sequenced is literally just garbage, dipped in amplicons still with the Primers in them to make a “fake” swab?
133. Why does the RaTG13 swab dated 27 November 2017 contain sequences more similar to SARS-CoV-2 than any other known virus?
134. Do Peter Daszak and WIV Scientists admit that TG13 or RaTG13 came from a degraded sample dipped in amplicons, enriched in primer dimers and tandem repeat products randomly ligated together by illumina preparation process, in an absence of any proper templates?
135. Do Peter Daszak and WIV scientists admit that it was impossible for the degraded RaTG13 sample to provide a proper "complete genome" by itself, and that fake NA sequence must have been produced by passage and spiked into the sample using viral amplicon products to make it look real?
Reference: https://ncbi.nlm.nih.gov/bioproject/PRJNA615303
Image 1:
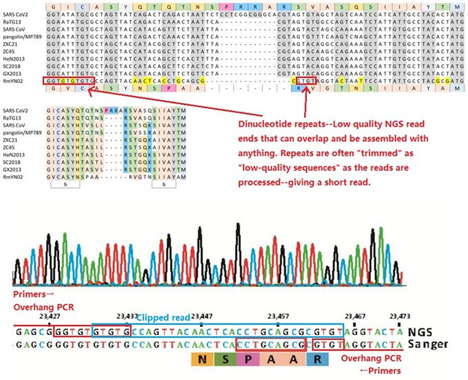
Image 2:
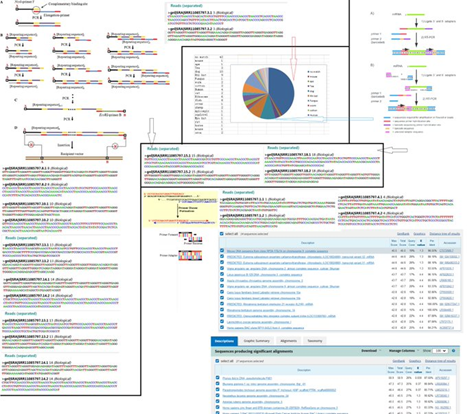
Image 3:

@AntGDuarte and @franciscodeasis
136. Does Peter Daszak in the statement found in the below image now admit that he was somehow involved in the Tongguan bat virus sampling expedition?

@AntGDuarte
137. How can you explain that the name of the virus (RaTG13) is on the SRA submitted to GenBank, and WIV researchers recently uploaded it on 2020-05-19, but as you can clearly see in the two images below, the files are dated 2018?
Image 1
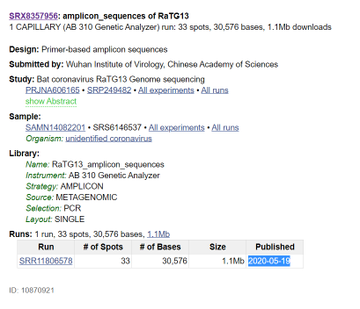
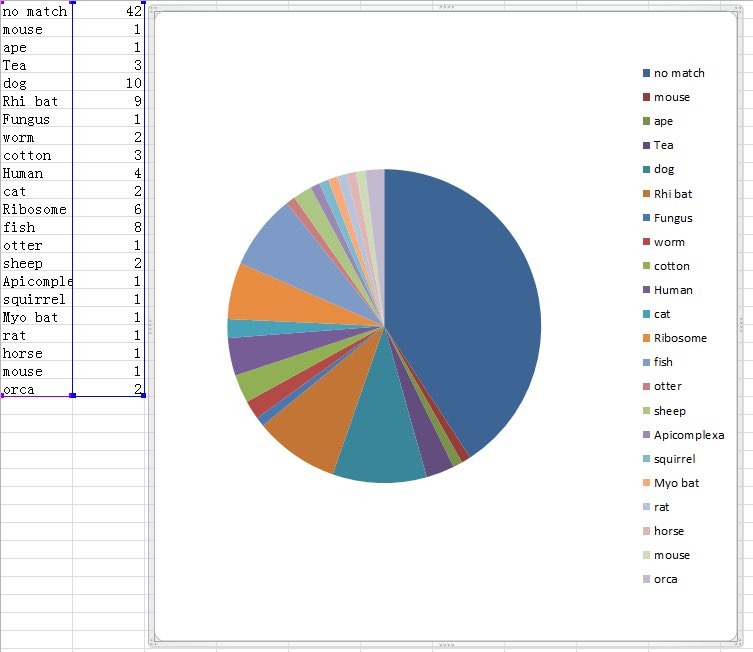
Image 2:
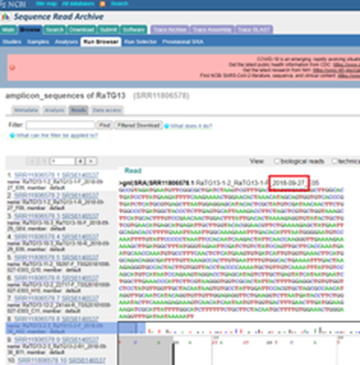
138. If, as evidenced above, WIV Researchers were studying RaTG13 in 2018, how can this fact be compatible with Petr Daszaks’s statements that: "We didn't do anything about it and put it in the freezer"?
@TheSeeker268
139. Will WIV and Peter Daszak go on the record to deny that WIV Scientists were studying BatCov/4991 and other as yet unpublished virus samples from Mojiang between 2013 and 2019?
Professor Zhengli Shi's team collected 97 samples from the Mojiang cave in 2013.
They were still studying those samples as late as December 2017
Reference: https://virosin.org/cn/article/doi/10.1007/s12250-018-0017-2
Image:
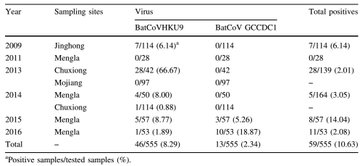
@franciscodeasis
140. Does this mean that 2017 was the year in which EcoHealthAlliance and Peter Daszak were finally allowed to see those samples from "that famous mineshaft"?
@interne41914499
141. What were the results of the 2019 MIST inspections and review of the WIV-led project 2013FY113500 that identified RaTG13?
142. Which virologists, medical and other associated staff members are currently under exit bans, unable to leave China?
143. Prof Zhang Yongzhen's team collected the first published SARS-CoV-2 sample as part of cooperation under the (no longer WIV-led) follow on MIST project. Which 50 researchers from which nine Chinese institutions were due to work on this project?
@franciscodeasis
144. Why there are dates from 2017 and 2018 in the filenames of the BatCOV/4991 reads
145. Peter Daszak facilitated a large upload of viral genome sequences to GenBank recently for IDs ranging 6233 to 9408 (2014-2015). Why has he not facilitated the uploading of previous missing IDs, from 2012 to 2013?
146. Why did WIV Bat research team not upload viral sequences from rodents and shrews from Yunnan? (Only Bat Researchers, Jin and Huang, declared that they had collected samples from them in Tongguan, although it was a normal practice in other locations (e.g. Fugong)
Via: Murat Seyran, email: muratseyran@gmail.com
https://vixra.org/abs/2005.0202
147. Why is the SARS-CoV-2 recombination pattern of the insert of furin cleavage site during host tropism unique and a onetime event which conflicts with the genomic composition of genetically related CoVs and template switching (copy-choice) mechanisms?
148. If SARS-CoV-2 genome is claimed to be almost identical to the origin Bat CoV, how do you explain that, it is only mutated on the RBD (another one-time event as well), the "too perfect" RBD does not have any high-frequency mutations in the clinical strains, and the RBD of SARS-CoV-2, unlike other human pathogenic CoVs, is not a positive selection site?
149. How did SARS-CoV-2 survive the immune system of putative pangolin intermediate hosts or even humans during the very first interactions with its flat easy-target sialic acid-binding domain?
150. SARS-CoV-2 displays a low frequency of mutations on its RBD, so how did the virus obtain such effective RBD compositions without being destroyed by pangolin or human immunity due to its easy target sialic acid-binding domain?
151. Why does only the RBD show mutations while the rest of the SARS-COV-2 genome remains almost unaltered?
152. Betacoronavirus lineage B CoVs including SARS-CoV S Protein does not have a pattern of recombination on S1/S2 region, so how did SARS-CoV-2 obtain that ability and why do we not see any further recombination in the clinical SARS-CoV-2 strains?
153. SARS-CoV-2 clinical strains do not have any high-frequency mutations on NTD and RBD, so how did the virus obtain such a perfect and precise host cell membrane interaction capacity? (Unlike SARS-Cov which perished due to its failed adaptation)
154. Why did this pandemic not emerge in places where people rely on water sources shared with bats or bats consumed as bush meat, but rather in Wuhan?
155. If SARS-CoV-2 is not a genetically engineered Bat CoV RaTG13, how do you explain its unnatural host tropism pattern and pandemic potential compared to other human pathogenic CoVs?
156. How do you explain that the host tropism and infection pattern of SARS-CoV-2 have 3 fundamental differences compared to the previous six human pathogenic CoVs: (HCoV-229E, HCoV-OC43, SARS-CoV, HCoV-HKU1, HCoV-NL63, and MERS-CoV)?
A. N-terminal domain (NTD) of CoVs Spike (S) Protein contains a specific glycan-binding region as the first contact area with the new host.
B. Specific glycan-binding immune receptors e.g. C-type lectins recognize NTD of S Protein of CoV and exterminate the virus before its adaptation.
C. According to the Canyon Hypothesis, CoVs sink this glycan-binding domain beneath the surface of S Protein to evade host immune system e.g. MERS-CoV glycan-binding domain 280 Å2 under its S Protein surface or HCoV-229E deleted its glycan-binding NTD during host tropism.
https://pubmed.ncbi.nlm.nih.gov/2670920/
and
https://www.jbc.org/content/264/25/14587.long
and
https://researchsquare.com/article/rs-16074/v1
Ref: C type letins interacts with SARS-CoV-2 Spike abundantly, many blocking ACE2 interaction.
157. Why does SARS-CoV-2 not have a single amino acid (aa.) alteration or deletion on its glycan-binding region NTD of its S Protein compared to its alleged parent virus BatCoV RaTG13?
158. How can you explain the fact that the flat and unsunken surface of SARS-CoV-2 NTD S Protein conflicts with the general adaptation and survival pattern of all previously known CoVs?
159. Based on the template-switching mode, CoVs pause their replication on certain domains and show recombination on these specific sites. However, other betacoronavirus lineage B members and the clinical strains of SARS-CoV-2 do not have any alterations on S Protein S1/S2.
SARS-CoV-2 and BatCoV RaTG13 are both betacoronavirus lineage B and their genomes are almost identical except 4 aa. Inserts between the S1/S2 which enables cleavage by host cell furin protease. What one-time unique event caused SARS-CoV-2 to obtain this trait?
160. After host adaptation, CoVs improve their host cell interaction with certain aa. substitutions on their receptor binding domain (RBD), considered as a positive selection site. SARS-CoV-2 has 22 aa. substitutions on S Protein RBD compare to BatCoV RaTG13. How do you explain as “natural” that, despite millions of SARS-CoV-2 infections, its RBD has not indicated a single high-frequency aa. substitution?
161. How do you explain as “natural” the completely unnatural and too perfect angiotensin converting enzyme 2 (ACE2) binding that was gained with a one-time alteration?
162. How do you explain as “natural” that, unlike the RBDs of other CoVs, SARS-CoV-2 RBD is not a positive selection site, indeed it has a flat and unaltered NTD, a conserved RBD and a unique S1/S2 insert of S Protein of SARS-CoV-2? This suggests an engineered “nature”.
163. Why is genetic engineering of CoVs considered a “conspiracy theory” and mere “speculation” when 18 research projects to develop genetically modify CoVs as pandemic potential pathogens were paused by United States Government Moratorium in 2014 and joint research with US gain of function virologists then continued in China?
@flavinkins
164. Why Zhengli Shi claim the discovery of RmYN02 containing a “inserted polybasic cleavage site nearly identical to CoV2” yet the actual sequence show only deletions and no polybasic cleavage site of any kind?

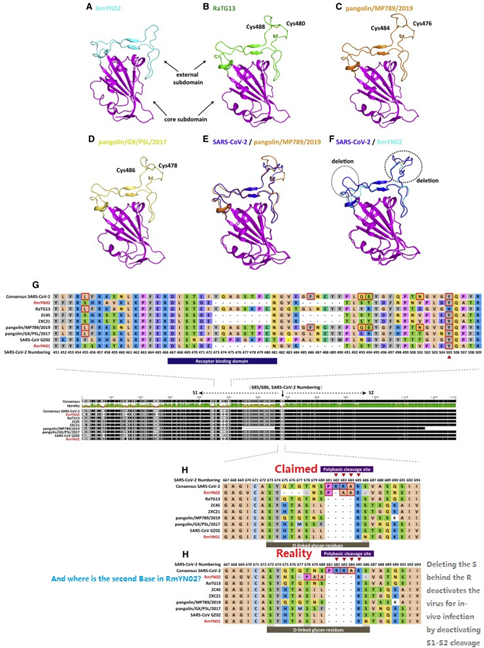
165. @flavinkins
http://whiov.cas.cn/tzgg_160286/202005/t20200511_5577837.html
At the critical dates when the SARS-CoV-2 genome was sequenced, many of the WIV's lab apparatuses were being disposed of.
What kind of the devices were and why were they disposed of?
https://news.sina.cn/gn/2020-03-10/detail-iimxyqvz9378276.d.html
166. How could such a RBD be acquired in the first place if pangolins are incapable of passing viruses to each other at all, hence will not be able to support such adaptation in the first place?
https://biorxiv.org/content/10.1101/2020.06.19.158717v1.full
via @Nomdeplumi1 and @franciscodeasis
167. What happened to the BALF/tissue/human samples they collected from miners?
(we know they collected these)
168. What is the long-term clinical course of the surviving miners who survived the first bought of Sars like Pneumonia back in 2012?
Are any still alive and most importantly do they still have the IgC of the Miners?

Why have images of their lungs not been shared?
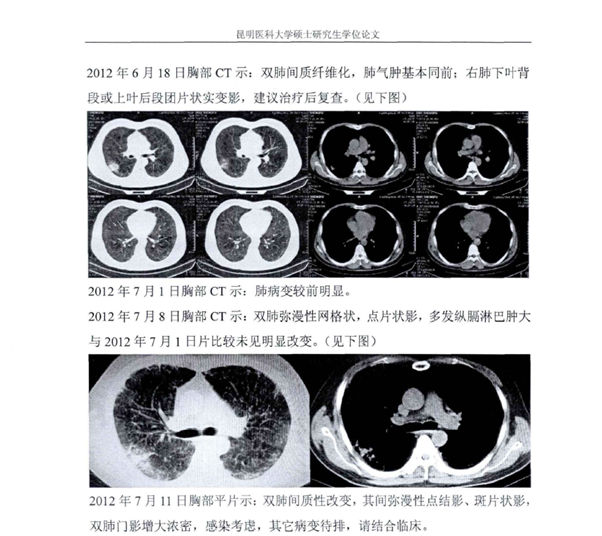
Via @billybostickson
169. Why did Andersen et al in their Nature letter claim predicted “O linked glycans” downstream of the furin cleavage site yet fail to retract their claims concerning a possible lab leak made on the basis of those predictions, when evidence has since emerged that their prediction was false:
O-linked glycans have been located at RBD T323/S325 on SI region arising from RBD mutation (not on RBM) linked to receptor binding & are not near S1-S2, clearly not "downstream from furin cleavage site"
Nor is it a “mucin-like domain” that stops immune recognition
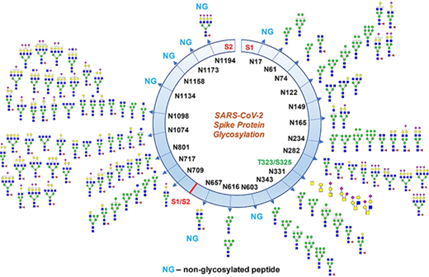
Via @AmaratungaGehan
170. Has any one from the WIV or WCDC reported or been treated for Covid 19 like symptoms before January 1st 2020, and if so when?
Via @liljurist
171. Who built and what were the design plans for WIV’s wastewater decontamination system?
Was there a final inspection before the lab was certified and operational?
(Has it been checked recently?)
171. From 2017-2019 was WIV receiving funding or researching a potential CoV or HIV vaccine?
172. To what extent did WIV share samples and collaborate with surrounding CDC labs, including animal testing?
173. How did Wang Yanyi become appointed director of WIV when she has very little relevant experience? Was it because of her relationship with her husband?
174. Was trypsin bought and stored at the lab? How much and when and for what was it used?
175. There seem to have been no significant publications by WIV Scientists since Zhengli Shi’s in January 2020. After SARS- CoV-2 was discovered, what has WIV been doing research into?
@billybostickson
176. Why have hundreds of potentially incriminatory website pages been deleted over the last few months on both the English and Chinese versions of the websites belonging to:
The Wuhan Insitute of Virology
Wuhan CDC
Wuhan University Institute of Model Animals
(Complete list is available on request)
177. Coming Soon!